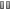| 03 Mars 2017
 (Okayama, 3 March) Researchers at Okayama University report in Scientific Reports how the overloading of protein transport mechanisms affects cell functioning. Over-expression of different types of target proteins leads to cellular growth defects, and the mechanism for transporting proteins out of a cell nucleus has the lowest over-expression tolerance.
(Okayama, 3 March) Researchers at Okayama University report in Scientific Reports how the overloading of protein transport mechanisms affects cell functioning. Over-expression of different types of target proteins leads to cellular growth defects, and the mechanism for transporting proteins out of a cell nucleus has the lowest over-expression tolerance.
Transporting proteins to where they need to be within an organism’s cell — protein targeting — costs resources. Too much expression (the synthesis and subsequent handling by the cell) of proteins that require transportation may limit the cell’s resources for moving other proteins around, potentially hampering proper cell functioning. A team of researchers from Okayama University led by Hisao Moriya has now investigated in detail what actually happens when protein-targeting resources become overloaded, and concluded that overloading situations lead to cellular growth defects.
For protein targeting, organisms rely on so-called signals: particular amino acid sequences within proteins that act as labels. Mitochondrial targeting signals (MTSs), signal sequences (SSs), nuclear localization signals (NLSs) and nuclear export signals (NESs) are important labels that drive proteins to mitochondria, to the endoplasmic reticulum, into the cell nucleus, and out of it, respectively. Moriya and colleagues attached signals of these types to green fluorescent proteins (GFPs) and, using a method they had developed before (‘genetic tug of war’), measured the expression limits of the modified GFPs.
By comparing results for signal-modified and unmodified GFPs, the researchers first observed that the added signals reduce expression limits of the modified GFPs — except for the NLS modification. NESs had the lowest limit; this is due to the exhaustion of a protein known as chromosomal maintenance 1 (Crm1) when overloading nuclear export processes mediated by Crm1. For MTS- and SS-modified GFPs, specific limiting factors could not be identified yet.
Moriya and colleagues assessed the physiological consequences of the reduced protein expression capabilities by gene analysis of the sets of messenger-RNA molecules (the transcriptomes) in the cells expressing modified GFP; they found that the artificially applied signals trigger various defects in cellular processes, similar to what happens with GFPs modified with a misfolding mutant.
The scientists also managed to obtain estimates for the critical amount of protein causing growth defects, via measurements of the number of modified GFPs produced. For MTS-, NES- and SS-modified GFPs the limits were 4%, 1% and 0.7%, respectively (the percentages are relative to the amount of unmodified GFP produced).
The findings of Moriya and colleagues show that the overloading of transport machineries in cells has various knock-on effects on cellular functioning. As the researchers point out: “understanding and controlling process overloads is beneficial for disease treatment and cellular engineering.”
Background
Genetic tug of war
Developed in the group of Hisao Moriya at Okayama University, the ‘genetic tug of war (gTOW)’ screening method enables estimating the over-expression limit of a particular protein. The method consists of first implanting the gene encoding the protein in yeast (Saccharomyces cerevisiae) cells, then generating copies of the gene and, finally, determining the copy-number limit; the latter serves as a proxy for the protein’s expression limit. The researchers used the gTOW technique for investigating the expression limit of various signal-modified green fluorescent proteins (GFPs).
Green fluorescent protein
Green fluorescent protein (GFP) displays green fluorescence upon exposure to blue-to-ultraviolet light. The GFP gene is often used in biomedical protein expression experiments. For instance, it has been shown that the GFP gene can be expressed in specific cells, particular organs, or whole organisms. Moriya and colleagues expressed GFPs modified with localization signals in yeast cells; they exploited the fluorescence property of GFPs for investigating the effects of the modifications on cells, including growth defects.

Caption
Representation of changes in gene expression levels (green: decreased, violet: increased) upon high-level expression of different types of modified green fluorescent proteins (GFPs). Clusters of genes with specific functional categories are marked by boxes; clearly, high-level expression of modified GFPs leads to defects in cellular functioning.
Reference
Reiko Kintaka, Koji Makanae, and Hisao Moriya. Cellular growth defects triggered by an overload of protein localization processes. Scientific Reports, 6, Article Number 31774, 2016.
DOI:10.1038/srep31774
http://www.nature.com/articles/srep31774
Reference (Okayama Univ. e-Bulletin): Associate Professor Moriya’s team
Correspondence to
Associate Professor Hisao Moriya, Ph.D.
Research Core for Interdisciplinary Sciences, Okayama University,
3-1-1 Tsushimanaka, Kita-ku, Okayama 700-8530, Japan
e-mail : Cette adresse email est protégée contre les robots des spammeurs, vous devez activer Javascript pour la voir.
http://tenure5.vbl.okayama-u.ac.jp/~hisaom/HMwiki/index.php?TopEnglish
Further information
Okayama University
1-1-1 Tsushima-naka , Kita-ku , Okayama 700-8530, Japan
Public Relations and Information Strategy
Website: http://www.okayama-u.ac.jp/index_e.html
Okayama Univ. e-Bulletin: http://www.okayama-u.ac.jp/user/kouhou/ebulletin/
Okayama Univ. e-Bulletin (PDF Issues): http://www.okayama-u.ac.jp/en/tp/cooperation/ebulletin.html
About Okayama University (You Tube):
https://www.youtube.com/watch?v=iDL1coqPRYI
Okayama University Image Movie (You Tube):
https://www.youtube.com/watch?v=_WnbJVk2elA
https://www.youtube.com/watch?v=KU3hOIXS5kk
Okayama University Medical Research Updates (OU-MRU)
http://www.okayama-u.ac.jp/eng/release/index_id210.html
Vol.2:Ensuring a cool recovery from cardiac arrest
Vol.3:Organ regeneration research leaps forward
Vol.4:Cardiac mechanosensitive integrator
Vol.5:Cell injections get to the heart of congenital defects
Vol.6:Fourth key molecule identified in bone development
Vol.7:Anticancer virus solution provides an alternative to surgery
Vol.8:Light-responsive dye stimulates sight in genetically blind patients
Vol.9:Diabetes drug helps towards immunity against cancer
Vol.10:Enzyme-inhibitors treat drug-resistant epilepsy
Vol.11:Compound-protein combination shows promise for arthritis treatment
Vol.12:Molecular features of the circadian clock system in fruit flies
Vol.13:Peptide directs artificial tissue growth
Vol.14:Simplified boron compound may treat brain tumours
Vol.15:Metamaterial absorbers for infrared inspection technologies
Vol.16:Epigenetics research traces how crickets restore lost limbs
Vol.17:Cell research shows pathway for suppressing hepatitis B virus
Vol.18:Therapeutic protein targets liver disease
Vol.19:Study links signalling protein to osteoarthritis
Vol.20:Lack of enzyme promotes fatty liver disease in thin patients
Vol.21:Combined gene transduction and light therapy targets gastric cancer
Vol.22:Medical supportive device for hemodialysis catheter puncture
Vol.23:Development of low cost oral inactivated vaccines for dysentery
Vol.24:Sticky molecules to tackle obesity and diabetes
Vol.25:Self-administered aroma foot massage may reduce symptoms of anxiety
Vol.26:Protein for preventing heart failure
Vol.27:Keeping cells in shape to fight sepsis
Vol.28:Viral-based therapy for bone cancer
Vol.29:Photoreactive compound allows protein synthesis control with light
Vol.30:Cancer stem cells’ role in tumor growth revealed
Vol.31:Prevention of RNA virus replication
Vol.32:Enzyme target for slowing bladder cancer invasion
Vol.33:Attacking tumors from the inside
Vol.34:Novel mouse model for studying pancreatic cancer
Vol.35:Potential cause of Lafora disease revealed
http://www.okayama-u.ac.jp/eng/research_highlights/index_id46.html
|
About Okayama University Okayama University is one of the largest comprehensive universities in Japan with roots going back to the Medical Training Place sponsored by the Lord of Okayama and established in 1870. Now with 1,300 faculty and 14,000 students, the University offers courses in specialties ranging from medicine and pharmacy to humanities and physical sciences. Okayama University is located in the heart of Japan approximately 3 hours west of Tokyo by Shinkansen. |